Turn Your Computer Into a Force for Scientific Discovery
How Folding@home Uses Idle PCs to Help Find Cures for Deadly Diseases
Imagine this: Your computer sits idle on your desk after a long day of emails, browsing, or gaming. But what if that unused power could be transformed into a tool against humanity’s deadliest diseases?
What if, right now, your laptop, workstation, or gaming rig could crunch complex biological simulations to help unlock cures for cancer, Alzheimer’s, Parkinson’s, HIV, COVID-19 variants, ALS, Huntington’s disease, and even rare genetic disorders?
That’s exactly what Folding@home makes possible.
What Is Folding@home?
Folding@home (FAH) is a distributed computing project that turns personal computers around the world into a massive virtual supercomputer. It’s run by a collaboration of scientists and institutions led by the Pande Lab at Stanford University (and later continued by Washington University in St. Louis).
The project’s goal is to simulate protein folding, molecular dynamics, and other biological processes that are crucial to understanding how diseases start and how we can stop them.
By contributing your computer’s spare CPU or GPU power, you help researchers perform experiments that would otherwise take years — or be impossible on a single supercomputer.
Why Protein Folding Matters
Proteins are made of long chains of amino acids that fold into intricate three-dimensional structures. This 3D shape determines what a protein can do — whether it builds tissues, repairs cells, or sends chemical signals.
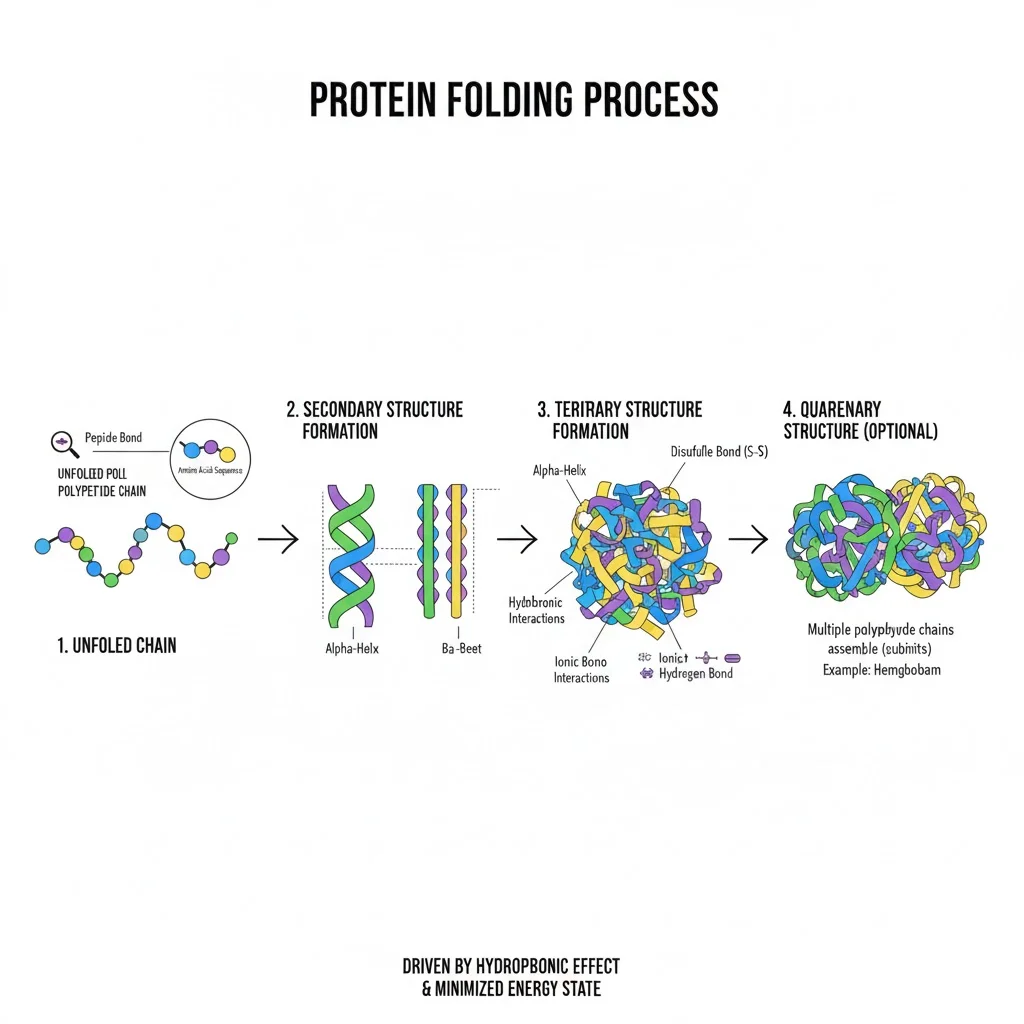
When proteins fold incorrectly, they can malfunction or clump together, leading to diseases such as:
- Alzheimer’s disease – caused by amyloid-beta protein misfolding
- Parkinson’s disease – linked to misfolded alpha-synuclein
- Cystic fibrosis – due to defective CFTR protein folding
- Cancer – from mutations that change protein shape or stability
Understanding how proteins fold — and what goes wrong when they don’t — allows scientists to design drugs that:
- Prevent misfolding
- Stabilize correct folding
- Block harmful interactions
- Promote protein repair or degradation
How Folding@home Works
Folding@home distributes extremely complex protein-folding simulations into small, manageable tasks called “work units.” Here’s what happens step by step:
1. Download and Install
You install the Folding@home client on your Windows, macOS, or Linux computer from the official website.
2. Connect and Run
The software connects to Folding@home servers and runs quietly in the background, using only your unused CPU/GPU resources.
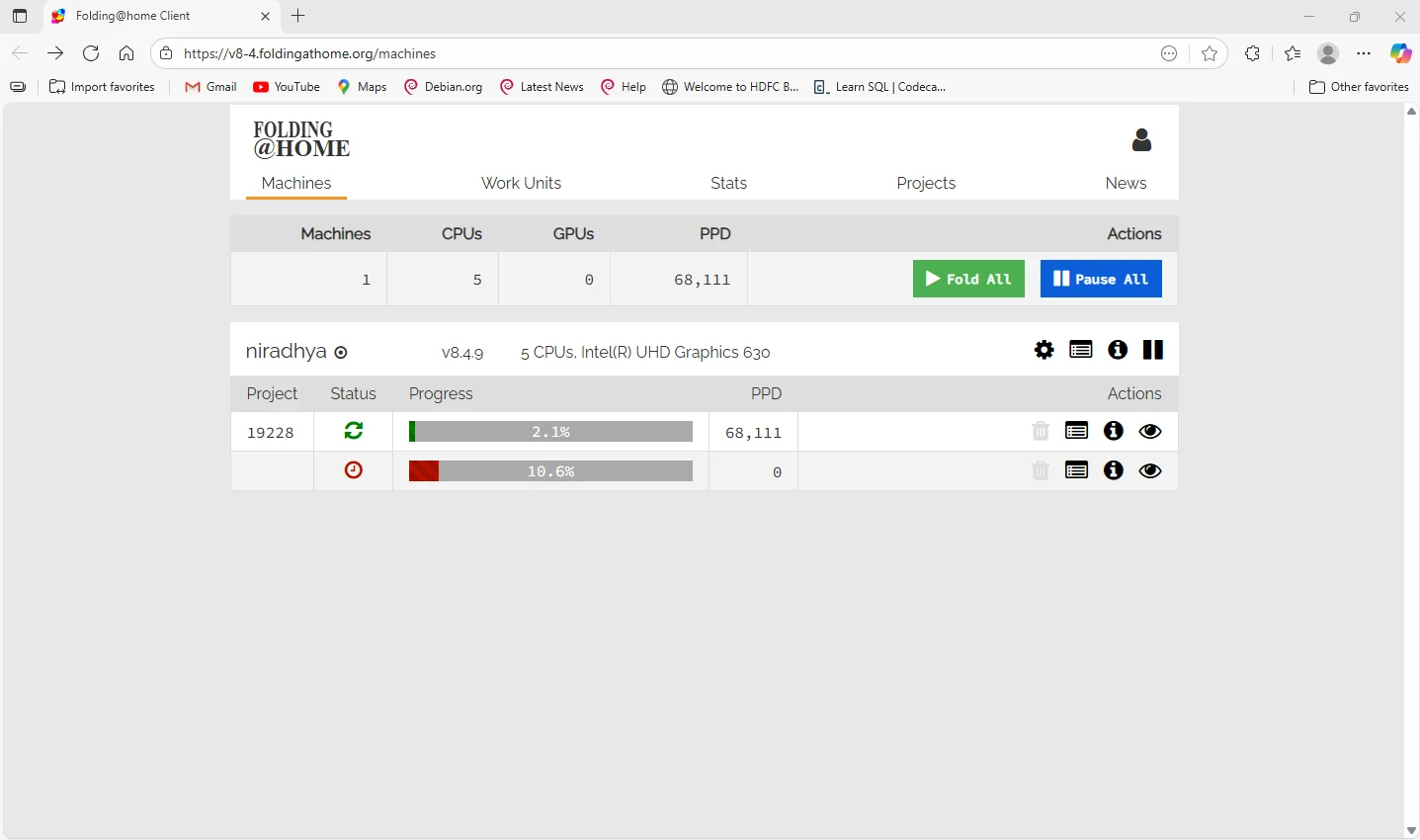
3. Process Work Units
Each work unit represents a small part of a large molecular simulation. Your computer processes one unit, then automatically uploads the results to the central servers.

4. Assemble and Analyze
Results from thousands of volunteers are combined to create a complete picture of how the protein moves, folds, or misfolds.
You can even join teams and track your collective progress through a point-based leaderboard system — adding a fun, competitive element to volunteering.
The Science Behind It
Simulating protein folding is one of the most demanding tasks in computational biology. Proteins constantly move, twist, and reshape themselves at lightning speed, and modeling every atom’s motion in real time requires massive computational power.
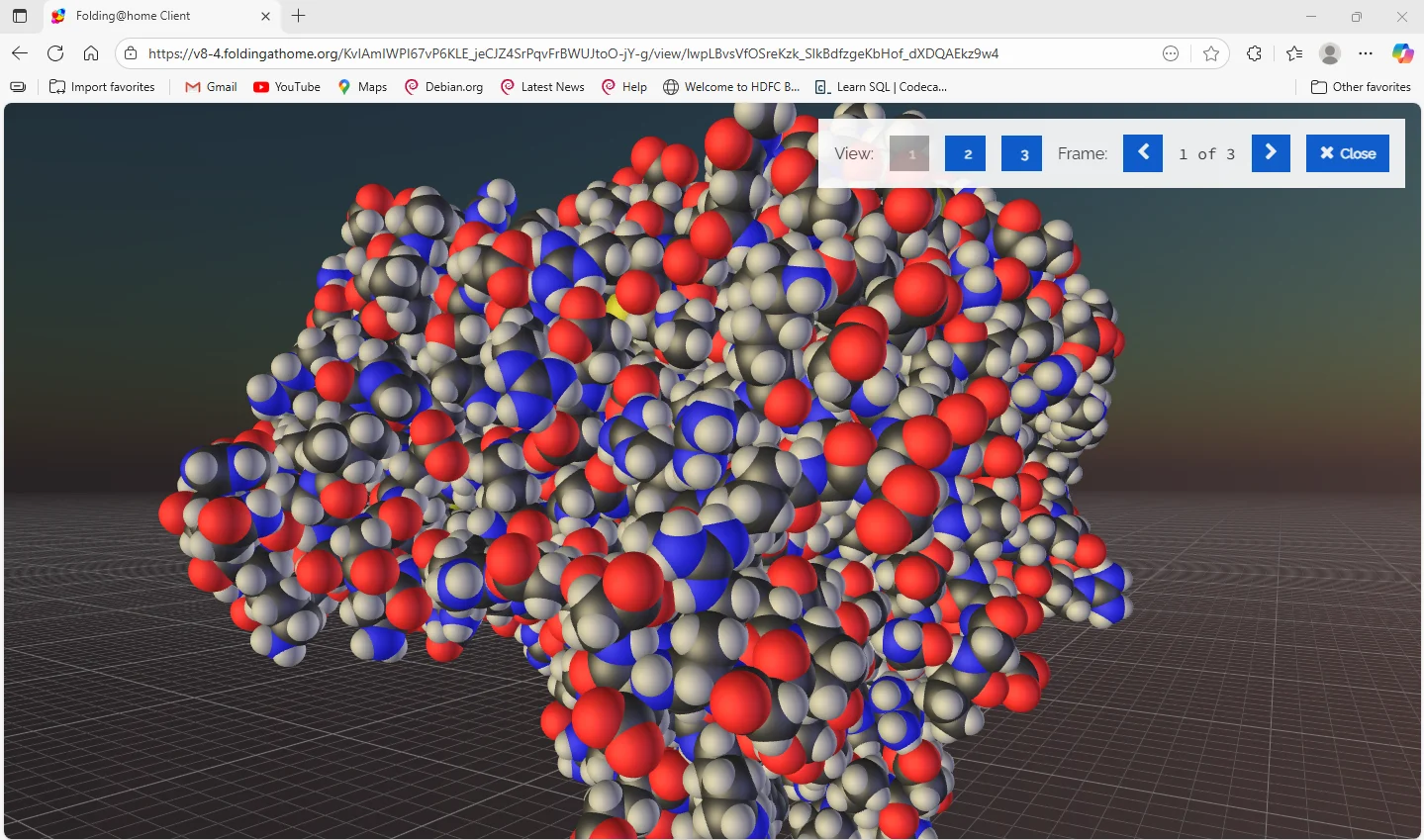
Instead of relying on one large supercomputer, Folding@home harnesses the power of hundreds of thousands of personal computers connected via the internet — together performing calculations faster than the top supercomputers in the world.
During the COVID-19 pandemic, Folding@home reached over 1 exaFLOP of computing power (that’s one quintillion operations per second!), making it the most powerful distributed supercomputer ever built.
Real-World Impact
Folding@home has contributed to research across a wide range of diseases. Projects have focused on:
- Cancer: Understanding how mutations destabilize key tumor-suppressor proteins.
- Alzheimer’s: Revealing pathways of amyloid-beta plaque formation.
- Parkinson’s: Identifying structures involved in toxic alpha-synuclein clumping.
- COVID-19: Mapping spike protein dynamics to support vaccine and antiviral development.
- HIV: Simulating viral protein interactions that drive infection.
- ALS & Huntington’s: Investigating aggregation mechanisms to guide therapies.
Why You Should Join
Running Folding@home doesn’t require any special skills or equipment — just a computer and an internet connection. Every volunteer contributes to a network that:
- Advances real biomedical research
- Supports universities and pharmaceutical scientists worldwide
- Demonstrates the power of citizen science and global collaboration
Your idle computer could help find the next breakthrough drug — or even a cure.